My research focuses on the ecology, diversity, and evolution of microorganisms and viruses in the environments. It studies a wide range of research subjects, spanning from genes to genomes, cells, ecosystems, and material cycles. My research methods encompass field sampling, cultivation, microscopy, and large-scale genome analysis. What makes microbial ecology fascinating is the vast spectrum of research scales in terms of both subject matter and methodology. This field is currently experiencing remarkable progress, thanks to the emergence of new approaches and technologies such as meta-omics and long-read sequencing.
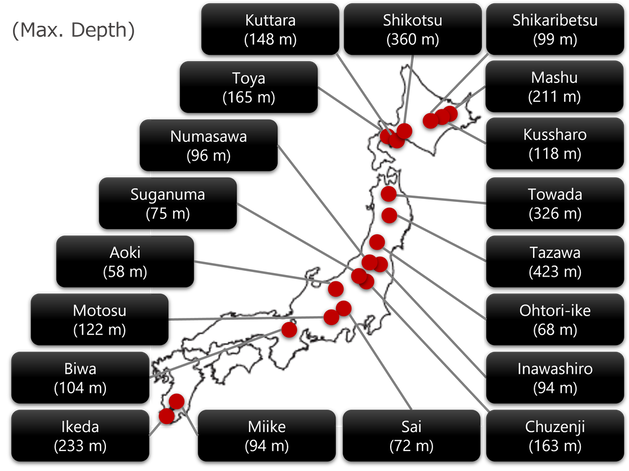
In particular, my research primarily revolves around deep freshwater lakes. It entails comprehensive work, from conducting field sampling to performing wet lab experiments and conducting bioinformatics analyses. Initially, my research focused on unraveling the diversity and ecological functions of lake microbial communies. However, I have recently developed an additional interest in leveraging the unique characteristics of lake ecosystems to gain a deeper understanding of microbial genomic and genetic diversity, and the driving forces behind their evolution.
My previous works
Visit here
My current research projects
Development of an ecogenomic database of lake microbial communities
(Mainly funded by JST Emergent Research Support Program)I am currently engaged in the development of a comprehensive environmental ecogenomic database. This project involves the collection of DNA and RNA samples from microbial eukaryotes, prokaryotes, and viruses in deep freshwater lakes across Japan. These samples are subsequently subjected to cutting-edge bioinformatics analyses, allowing for multifaceted investigations into the diversity, ecology, and evolution of environmental microorganisms. In particular, I consider each lake as a "replicate" or "control" of an ecosystem and performing a high-resolution inter-lake comparison of microbial diversity. By undertaking this endeavor, I aim to elucidate the underlying mechanisms responsible for the generation and maintenance of microbial genome and genetic diversity.
Challenging in isolating hypolimnion-specifc bacterioplankton lineages
(Mainly funded by Kyoto University's 125th Anniversary Fund "Kusunoki 125")Previous studies have revealed the presence of unique microbial communities and ecosystems in deep freshwater systems. To gain a deeper understanding of their ecology, researchers have employed culture-independent approaches such as FISH, 16S rRNA gene amplicon sequencing, and metagenomic analysis. However, the physiological and ecological characteristics inferred from these indirect methods remain speculative, and it is desirable to experimentally verify them through their isolation. By obtaining isolated cultures, I can not only validate the hypotheses but also investigate the responses of these microorganisms to various environmental stresses, and even isolate their associated viruses. In my ongoing research, I am undertaking the challenge of isolating bacteria that are typically difficult to culture in lakes, particularly those belonging to hypolimnion-specific lineages. To achieve this, I am utilizing high-throughput dilution-to-extinction cultivation, a method that has successfully isolated numerous oligotrophic bacterioplankton lineages so far.
Comprehensive identification of virus-host pairs in lakes using single cell genomics
(Mainly funded by JSPS KANENHI)Previous studies have extensively gathered genomic information on bacteria and their viral (phages) in lake water. However, a significant challenge remains in linking these genomes together to identify virus-host pairs. Without this crucial information, the ecological significance of each viral lineage remains elusive. In collaboration with Takeyama Laboratory at Waseda University, I am undertaking the task of identifying virus-host pairs in a non-selective and comprehensive manner, by employing single-cell genomics technology and detecting viral genome fragments within individual bacterial (host) cells. Furthermore, single-cell analysis provides the opportunity to recover highly diverse genomic regions that are challenging to assemble using metagenomic approaches. Leveraging this advantage, I am striving to elucidate the genomic diversity within a population at an unprecedented resolution, surpassing the limitations of metagenomics.
Other colloborative researches
Based on the extensive data and samples I have collected from numerous lakes, as well as my expertise and experience in this field, I am engaged in collaborations with researchers from diverse research fields. In addition to the works that have already been published as papers, for instencem, I am now involved in genome analysis of eukaryotic microorganisms and giant viruses present in lakes, as well as exploring biogeochemical aspects related to microbial systems. If you have any interest or ideas regarding the utilization of the lake microbial samples and genomic data, please do not hesitate to contact me.